Improved enzymes with new functions
Directed evolution: the most revolutionary protein engineering method known to date
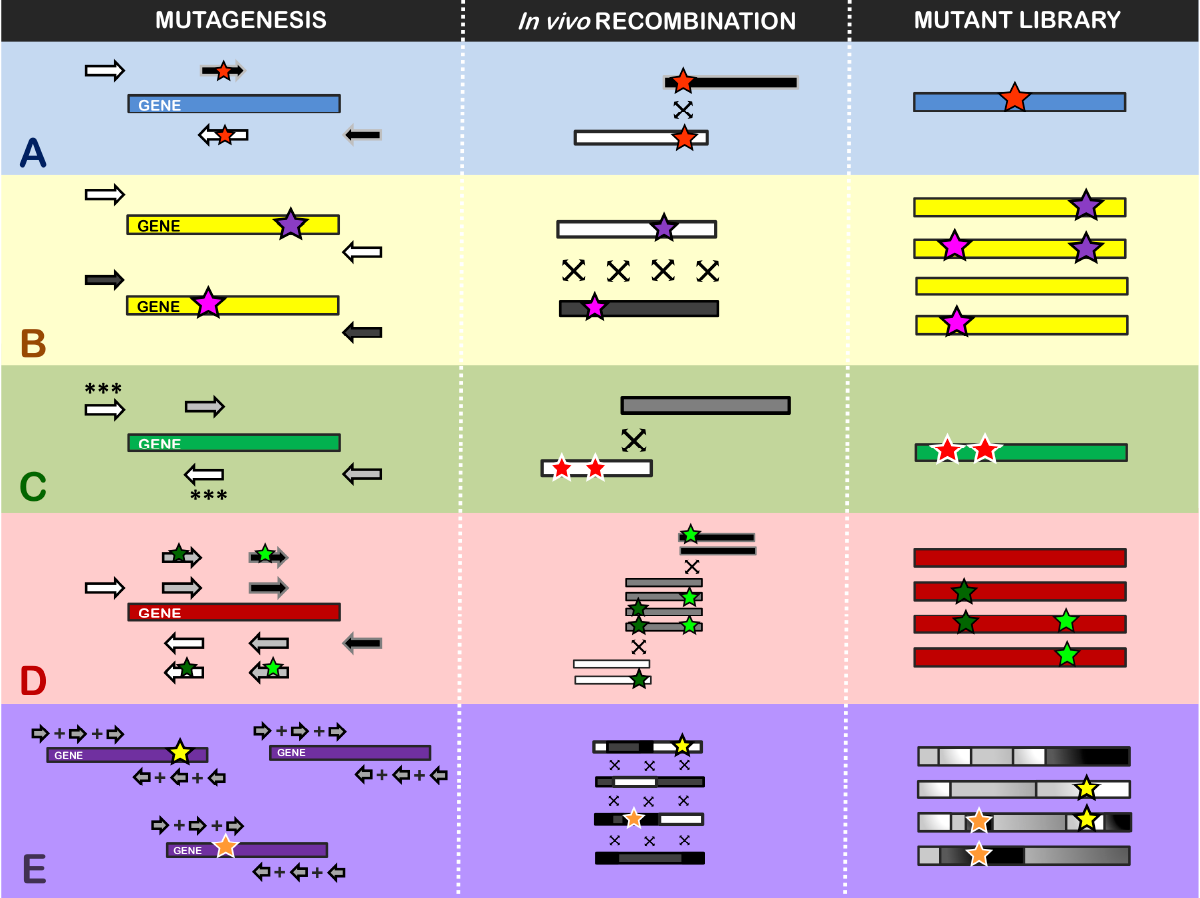
We use directed evolution to customized enzymes for different purposes and applications. In this procedure, we first create genetic diversity by random mutagenesis and/or the recombination of parental genes.
Once the mutant libraries are expressed in a suitable host organism, we can screen them in high-throughput platforms to search for specific biochemical traits. These include activity under extreme pH or temperatures, changes in substrate preferences, tolerance in the presence of strong inhibitors…
In this way, we can strictly control selective pressure until the desired properties are attained.
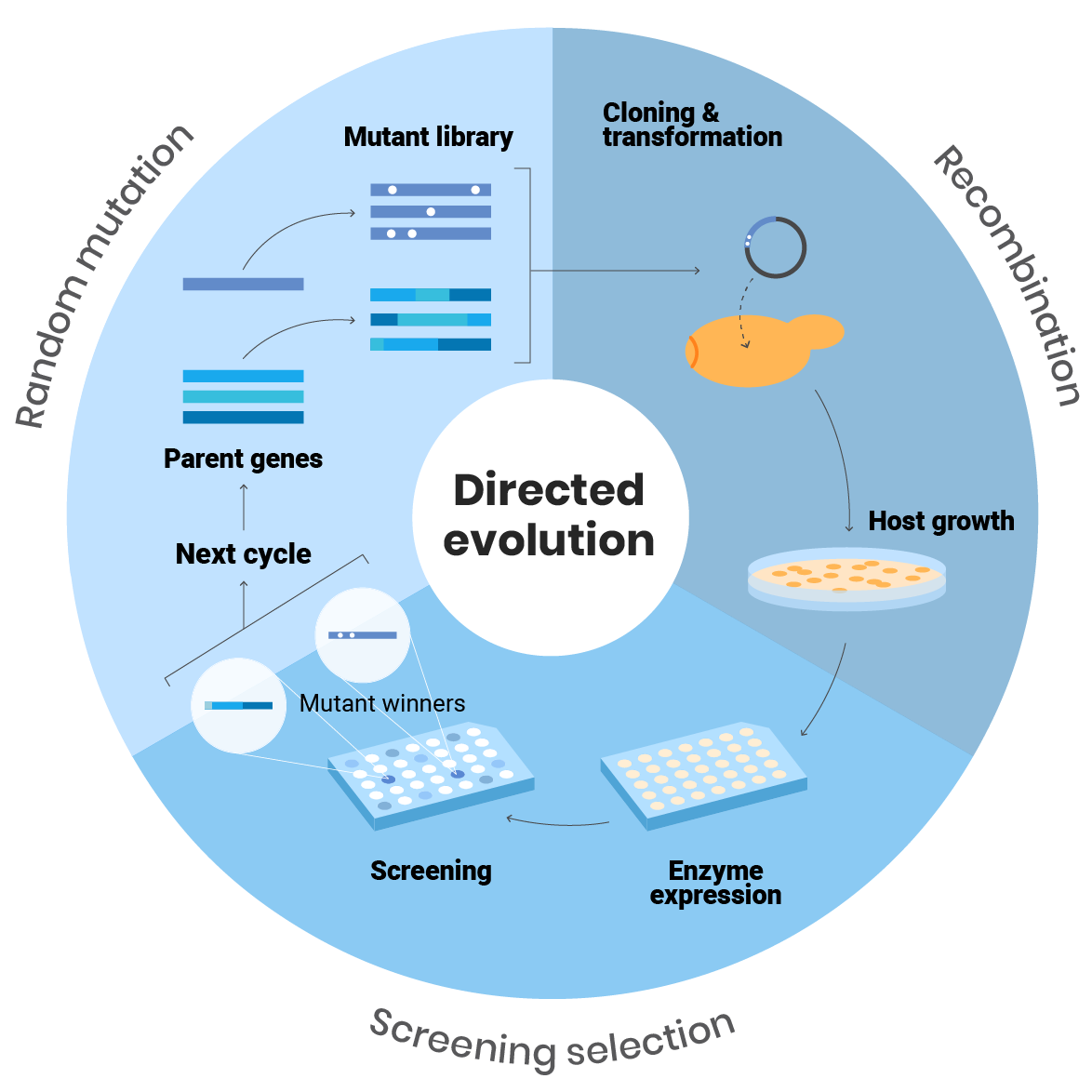
Leveraging Nature’s Blueprint
The Impact of Directed Evolution in Protein Engineering, an interview with Miguel Alcalde
Our Co Funder & Advisor Miguel Alcalde was interviewed by Azo Life Sciences at SLAS EU 2023 in Brussels, Belgium. Watch a fragment of the interview here or read the whole article.
Increased yield and efficiency of industrial processes
Our technological platform EVOSHUFFLER takes biocatalysts to the next level
With an ensemble of top-notch technologies in directed evolution based on the Saccharomyces cerevisiae machinery, we harness a variety of methods aimed at designing customized biocatalysts.
This unique approach offers huge versatility and ad-hoc solutions to different sectors, including the pharmaceutical, chemical, energy and environmental industries.
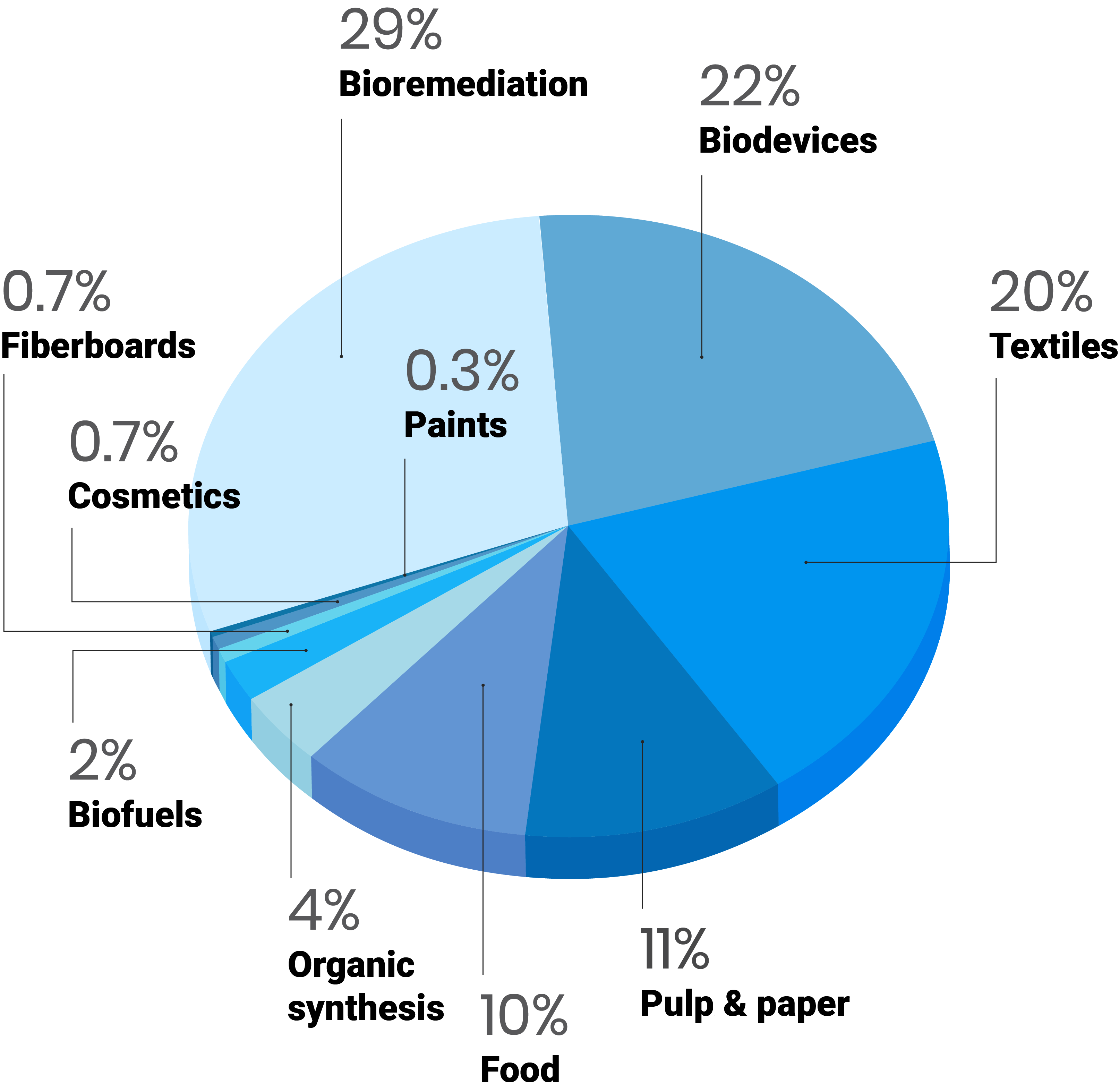
Patents
Unspecific peroxygenase mutants with high monooxygenase activity and their applications.
Molina-Espeja, P., Plou, F.J. Gomez de Santos, P., and Alcalde, M.
Method to degrade Plastic.
González Barroso, M.M., Torres Salas, P., Espi Guzman, E.,Gómez Fernández, B.J., Viña González, J., Mateljak, I.
Publications
Get to know the latest work from our team

The colors of peroxygenase activity: Colorimetric high-throughput screening assays for directed evolution
Dolz, M., Monterrey, D.T., Beltrán-Nogal, A., Menés-Rubio, A., Keser, M., González-Pérez, D., Gómez de Santos, P., Viña-González, J. and Alcalde, M. (2023) The Colors of Peroxygenase Activity: Colorimetric High-Throughput Screening Assays for Directed Evolution.
Methods in Enzymology: P450 enzymes. Renata, H. Ed. Elsevier.Vol 693: 73-109.
https://doi.org/10.1016/bs.mie.2023.09.006.
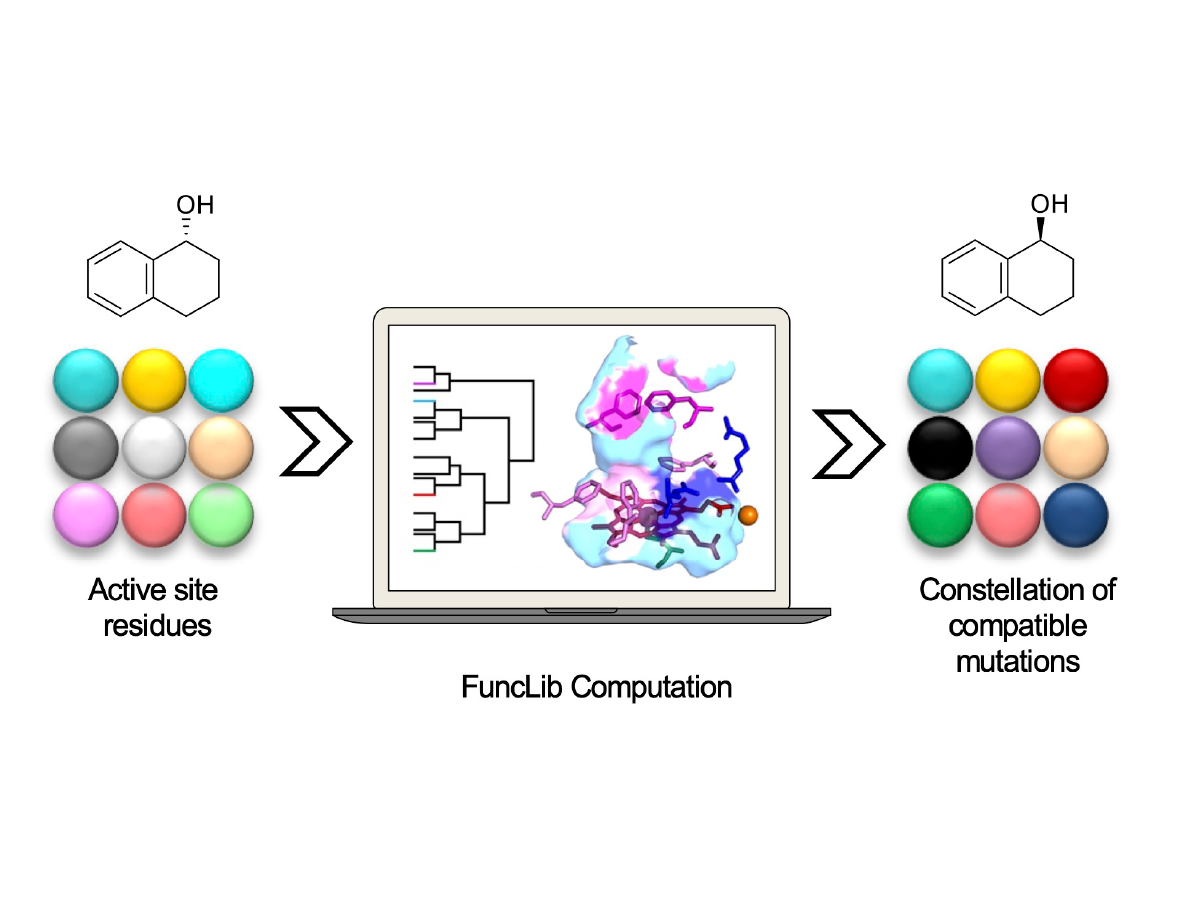
Repertoire of computationally designed peroxygenases for enantiodivergent C−H oxyfunctionalization reactions.
Gomez de Santos, P., Mateljak, I., Hoang, M.D., Fleishmann, S.J., Hollmann, F. and Alcalde, M. (2023).
Journal of the American Chemical Society. In press. https://doi.org/10.1021/jacs.2c11118.
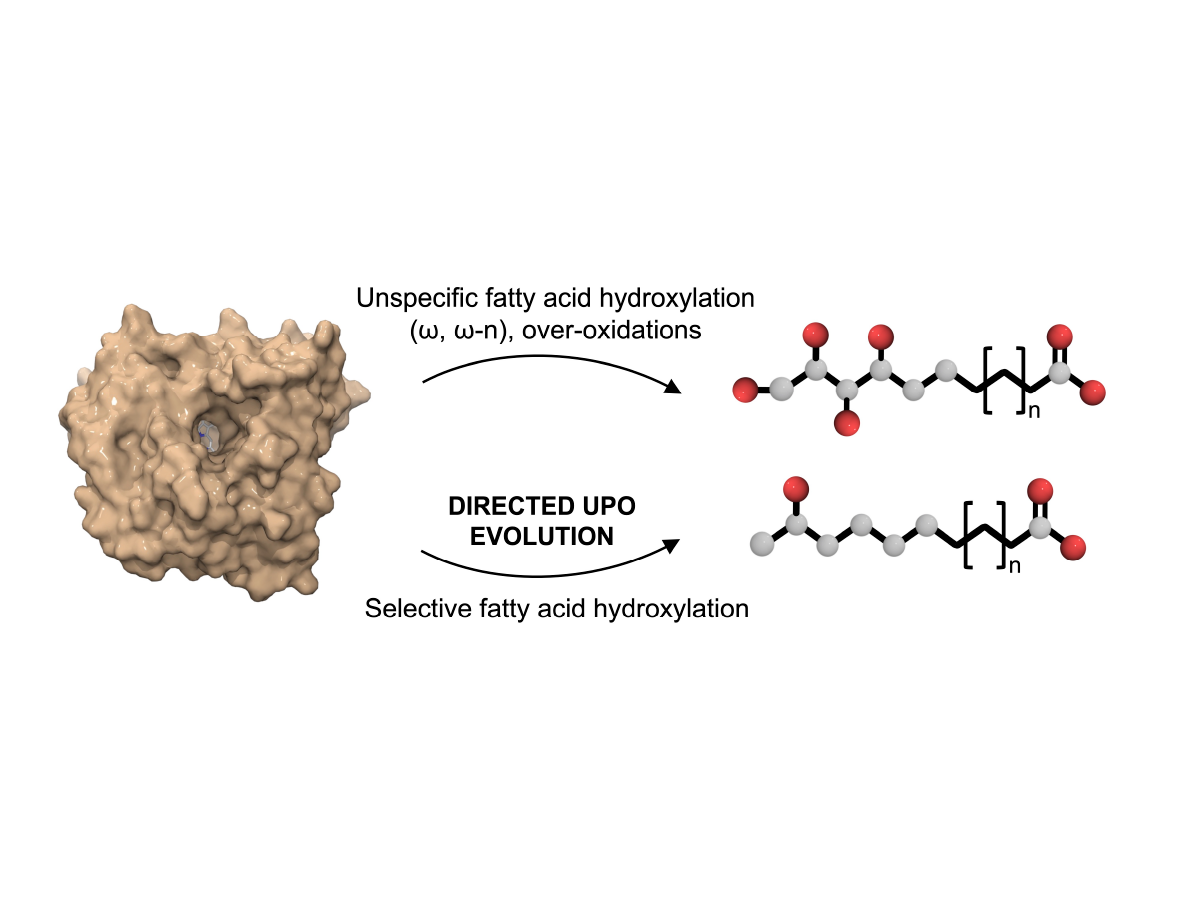
Engineering a highly regioselective fungal peroxygenase for the synthesis of hydroxy fatty acids.
Gomez de Santos, P., Gonzalez-Benjumea, A., Fernandez-Garcia, A., Aranda, C., Wu, Y., But, A., Molina-Espeja, P., Mate, D., Gonzalez-Perez, D., Zhang, W., Kiebist, J., Scheibner, K., Hofrichter, M., Swiderek, K., Moliner, V., Sanz-Aparicio, J., Hollmann, F., Gutierrez, A. and Alcalde, M. (2022).(2022).
Angewandte Chemie International Edition. e202217372.https://doi.org/10.1002/anie.202217372.
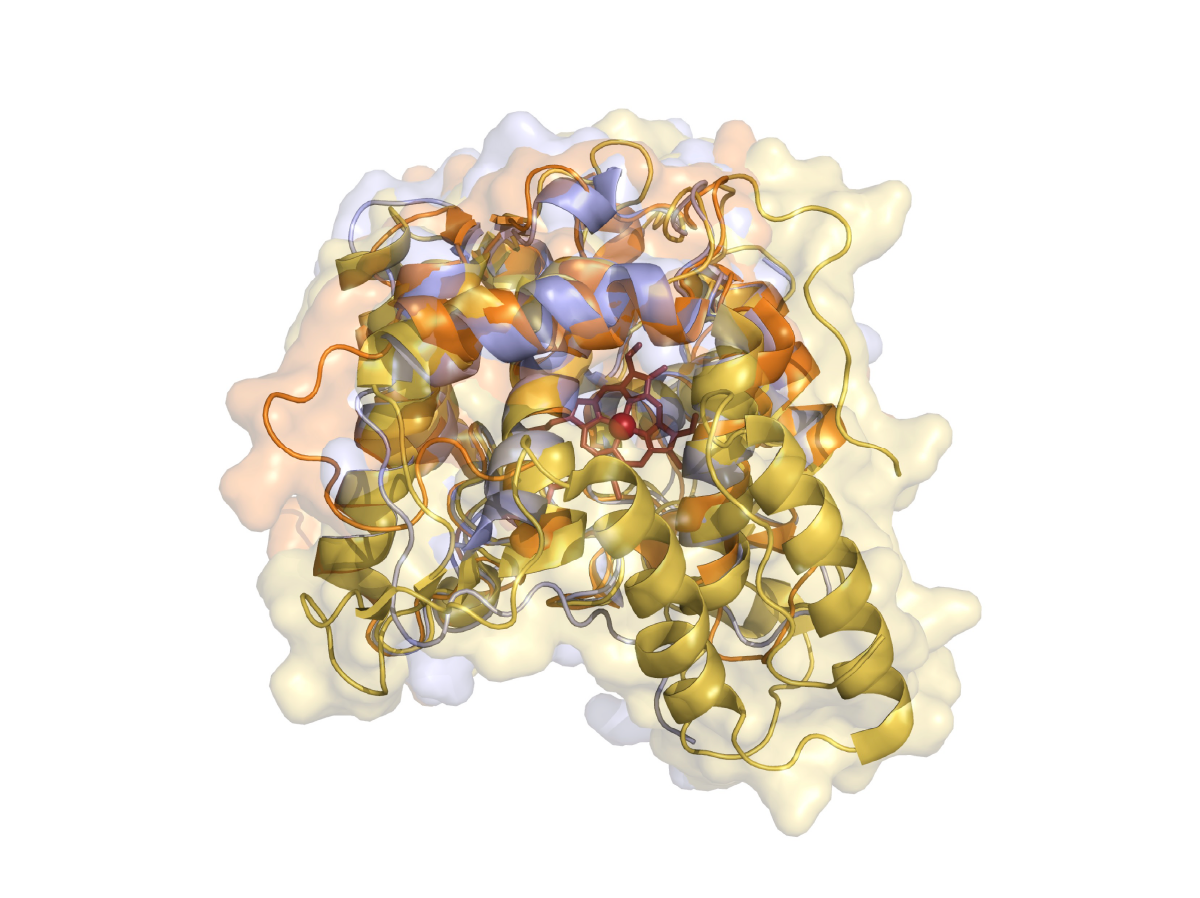
Surfing the wave of oxyfunctionalization chemistry by engineering fungal unspecific peroxygenases.
Beltran-Nogal, A., Sanchez-Moreno, I., Mendez-Sanchez, D., Gomez de Santos, P., Hollmann, F. and Alcalde, M. (2022).
Current Opinion in Structural Biology 73: 102342https://doi.org/10.1016/j.sbi.2022.102342.
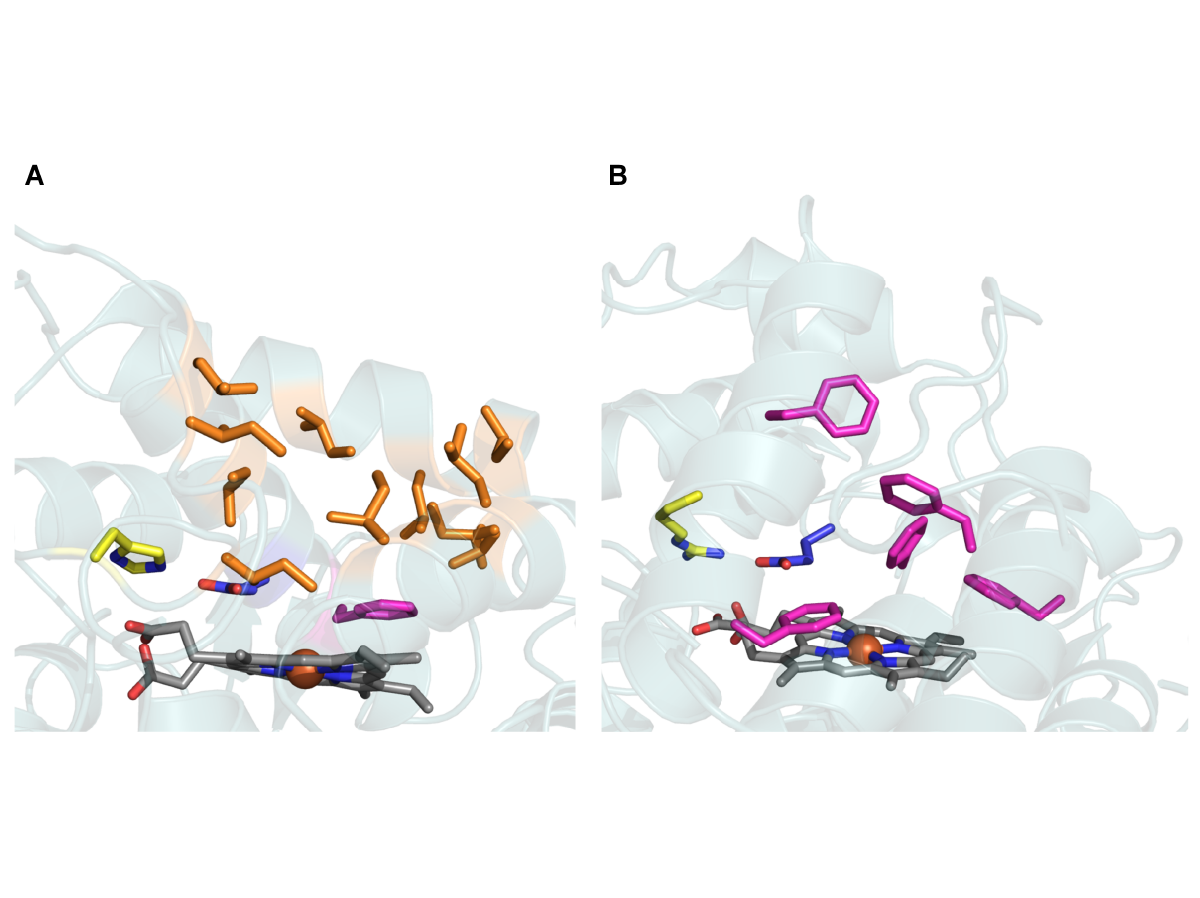
Directing the evolution of the fungal ligninolytic secretome.
Viña-Gonzalez, J. and Alcalde, M. (2021).
In: Protein Engineering: Tools and Applications. Zhao, H., Lee, S.Y., Nielsen, J. Stephanopoulos, G. Ed. Wiley-VCH, Weinheim, Germany. 295-311. ISBN 978-3-527-34470-3.https://doi.org/10.1002/9783527815128.ch12.
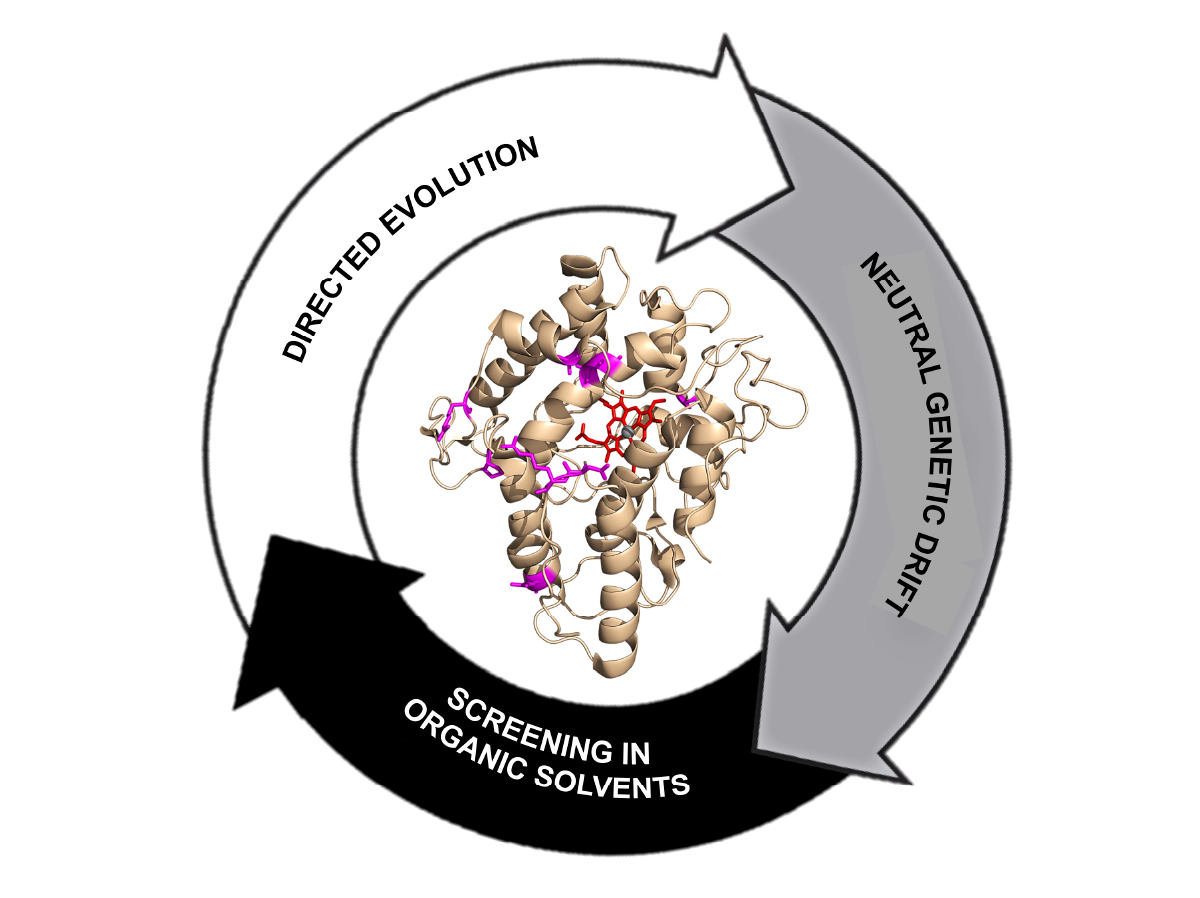
Directed evolution of the unspecific peroxygenase from Agrocybe aegerita in organic solvents.
Martin-Diaz, J., Molina-Espeja, M., Hofrichter, M., Hollman, F. and Alcalde, M. (2021).
Biotechnology & Bioengineering 118: 3002-3014https://doi.org/10.1002/bit.27810
Recent developments in the use of peroxygenases – exploring their high potential in selective oxyfunctionalisations.
Hobisch, M., Holtmann, D., Gomez de Santos, P., Alcalde, M., Hollmann, F. and Kara, S. (2021).
Biotechnology Advances 51: 107615. https://doi.org/10.1016/j.biotechadv.2020.107615
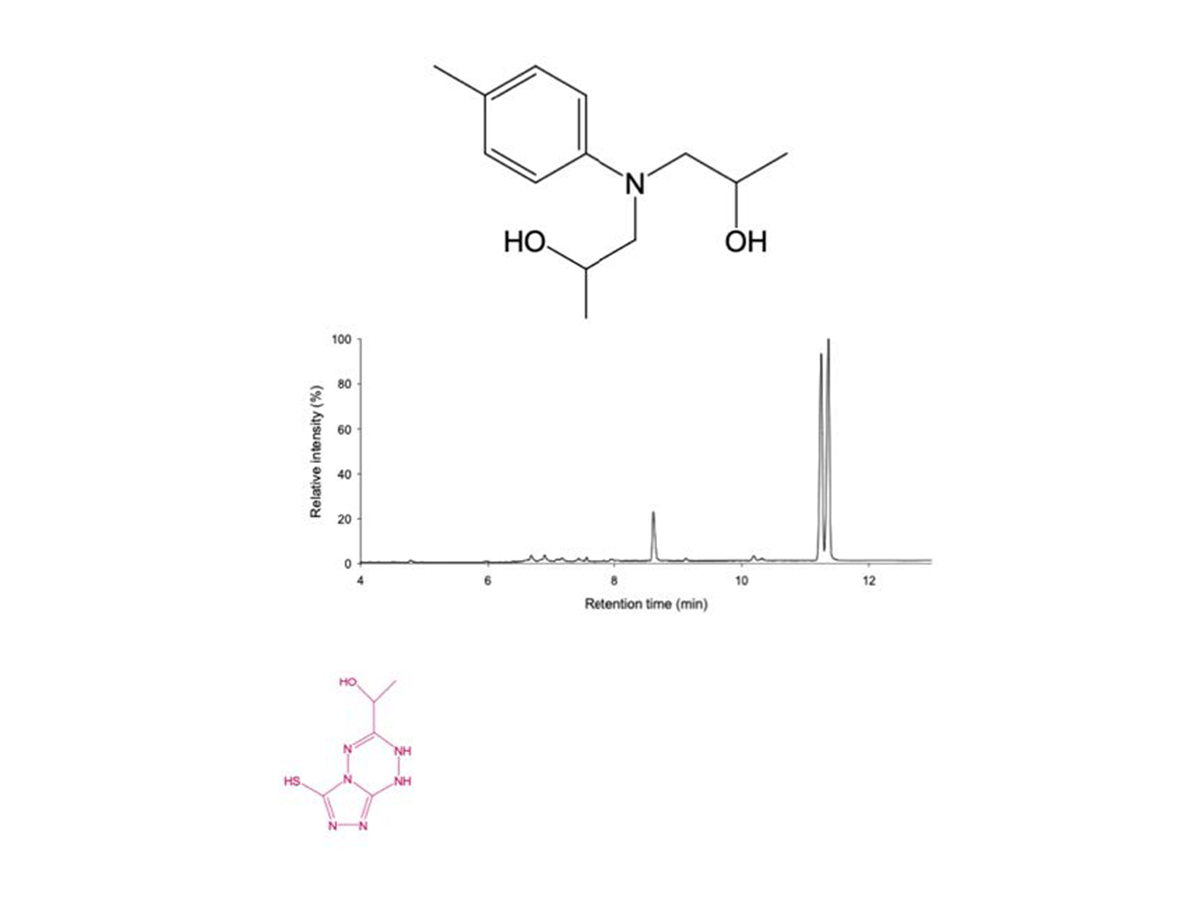
Colorimetric high-throughput screening assay to engineer fungal peroxygenases for the degradation of thermoset composite epoxy resins.
Dolz, M., Mateljak, I., Sanchez-Moreno, I., Mendez-Sanchez, D., Viña-Gonzalez, J. and
Alcalde, M. (2022).
Frontiers in Catalysis 2: 883263. https://doi.org/10.3389/fctls.2022.883263.
Engineering a Highly Thermostable High-Redox Potential Lacasse.
Mateljak I, and Alcalde M (2021)
ACS Sustainable Chem. Eng. 2021, 9, 29, 9632–9637 https://pubs.acs.org/doi/10.1021/
acssynbio.8b00509.

Functional Expression of Two Unusual Acidic Peroxygenases from Candolleomyces aberdarensis in Yeasts by Adopting Evolved Secretion Mutations.
Gómez de Santos, P; Hoang, MD; Kiebist, J; Kellner, H; Ullrich, R; Scheibner, K; Hofrichter, M; Liers, C and Alcalde, M (2021)
Applied and Environmental Microbiology 87 (19): e00878-21 https://pubs.acs.org/doi/10.1021/acssynbio.8b00509.
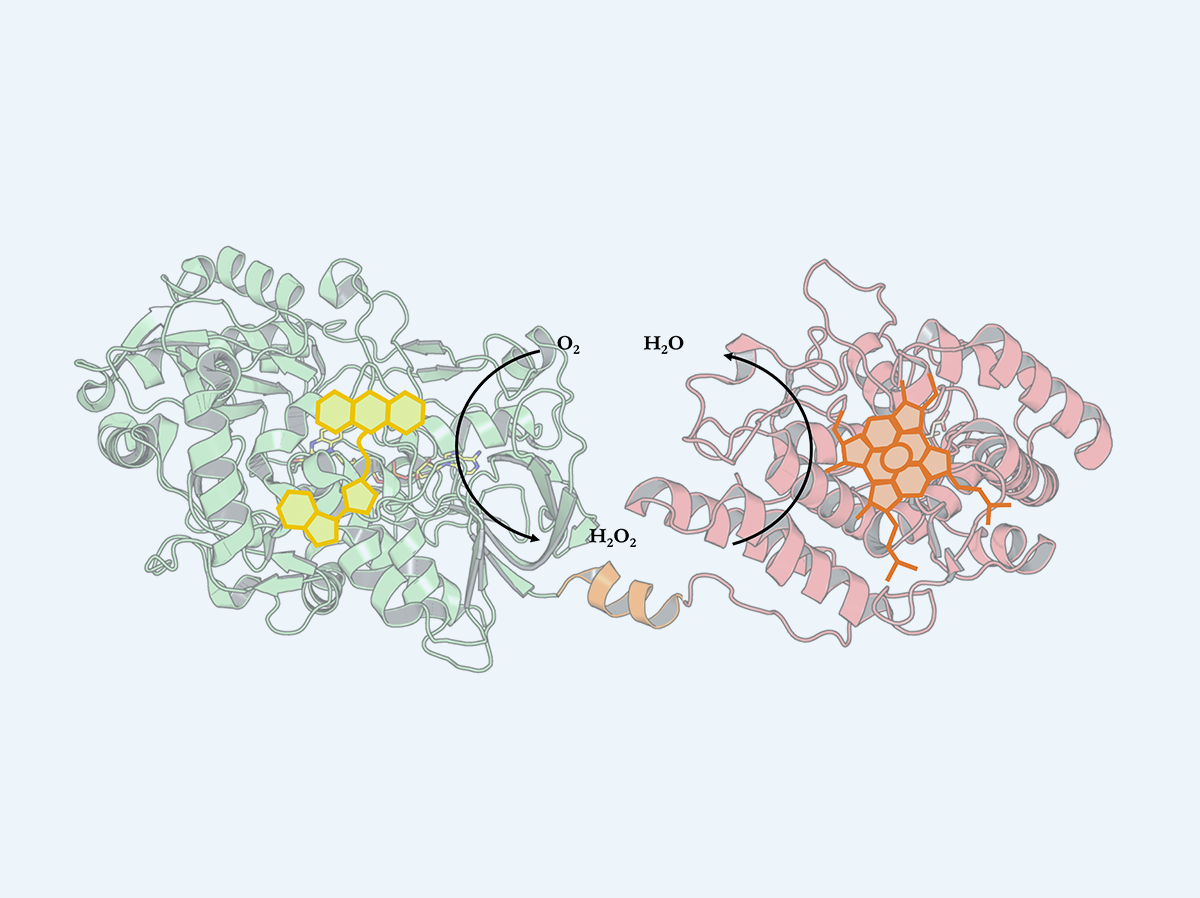
Evolved peroxygenase-aryl alcohol oxidase fusions for self-sufficient oxyfunctionalization reactions.
Gómez-Fernández, B.J., Risso, V.A., Rueda, A., Sánchez-Ruiz, J.M. and Alcalde, M.(2020).
ACS Catalysis 10: 13524-13534. https://pubs.acs.org/doi/10.1021/acscatal.0c03029

Ancestral Resurrection and Directed Evolution of Fungal Mesozoic Laccases.
Gómez-Fernández, B.J., Risso, V.A., Rueda, A., Sánchez-Ruiz, J.M. and Alcalde, M.(2020).
Applied and Environmental Microbiology 86 (14): e00778-20. JOURNAL COVER. https://pubs.acs.org/doi/10.1021/acssynbio.8b00509.
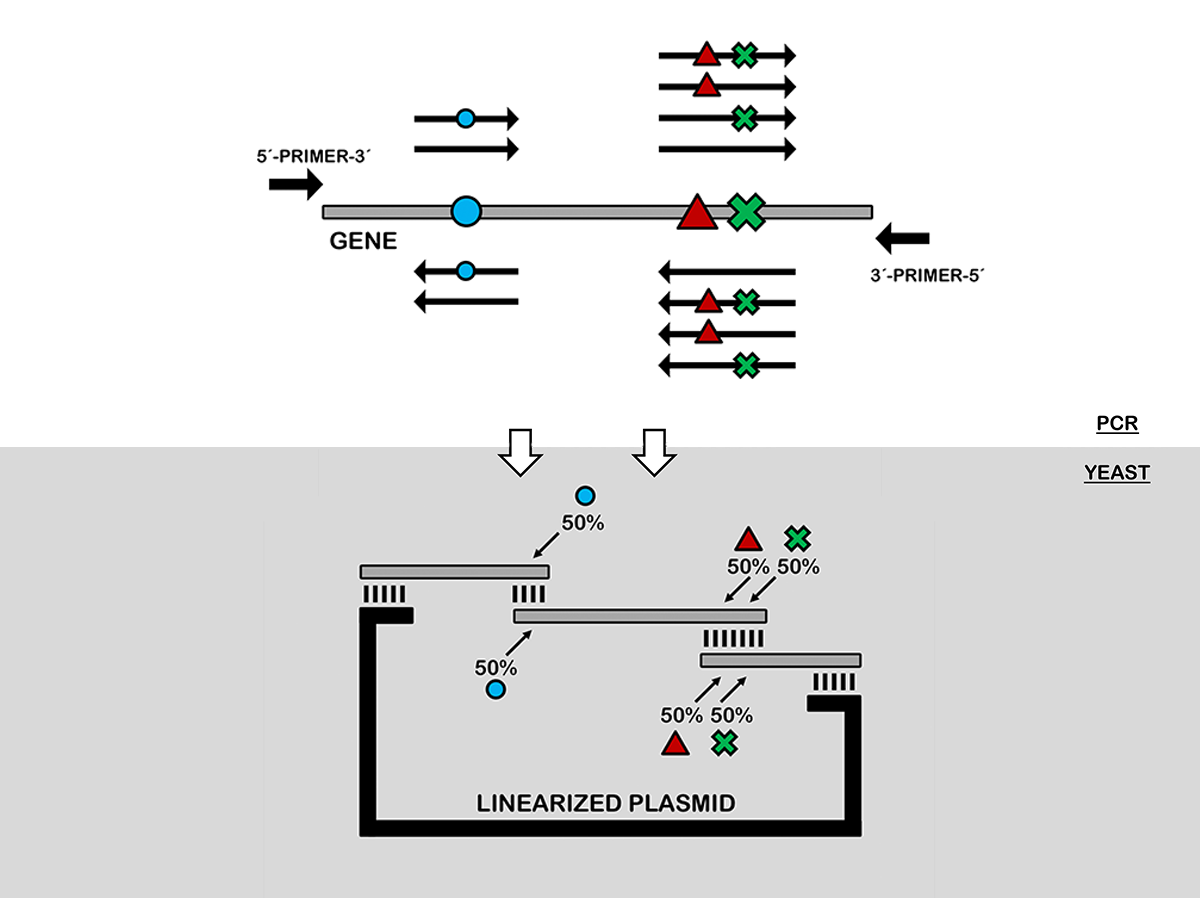
In vivo site-directed recombination (SDR): An efficient tool to reveal beneficial epistasis. In: Methods in Enzymology 643.
Viña-Gonzalez, J. And Alcalde, M. (2020).
Tawfik, D. Ed. Academic Press Elsevier Inc, Cambridge, MA, EEUU. Pages 1-12. ISBN 978-0-12-821149-6. https://pubs.acs.org/doi/10.1021/acssynbio.8b00509.
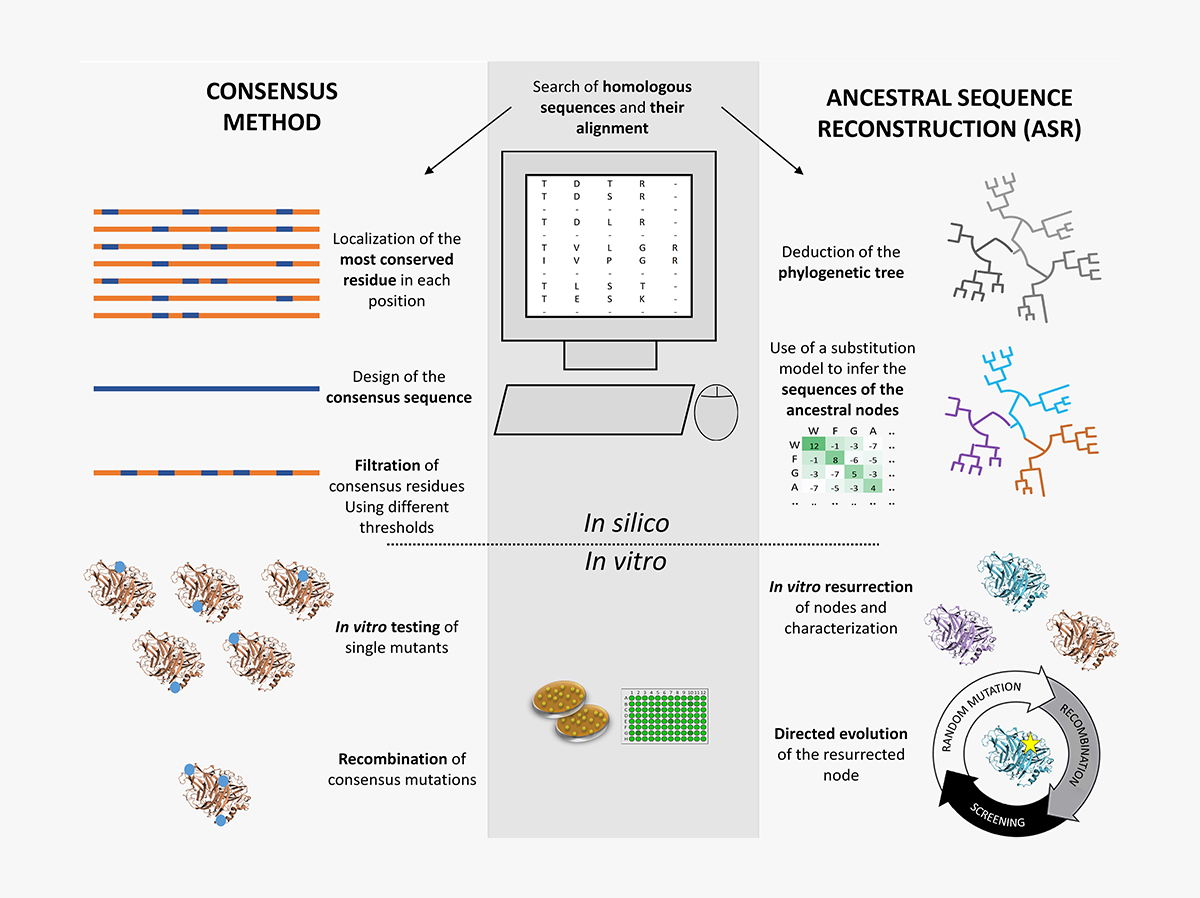
Laccase engineering by directed and computational evolution. In: Laccase in bioremediation of pollutants and xenobiotics.
Mateljak, I., Gomez-Fernandez, B. and Alcalde, M. (2020).
Bioremediation and Waste valorization. Schlosser, D., Ed. Springer Nature Switzerland. Pages: 191-206. ISBN: 978-3-030-47906-0 https://pubs.acs.org/doi/10.1021/acssynbio.8b00509.
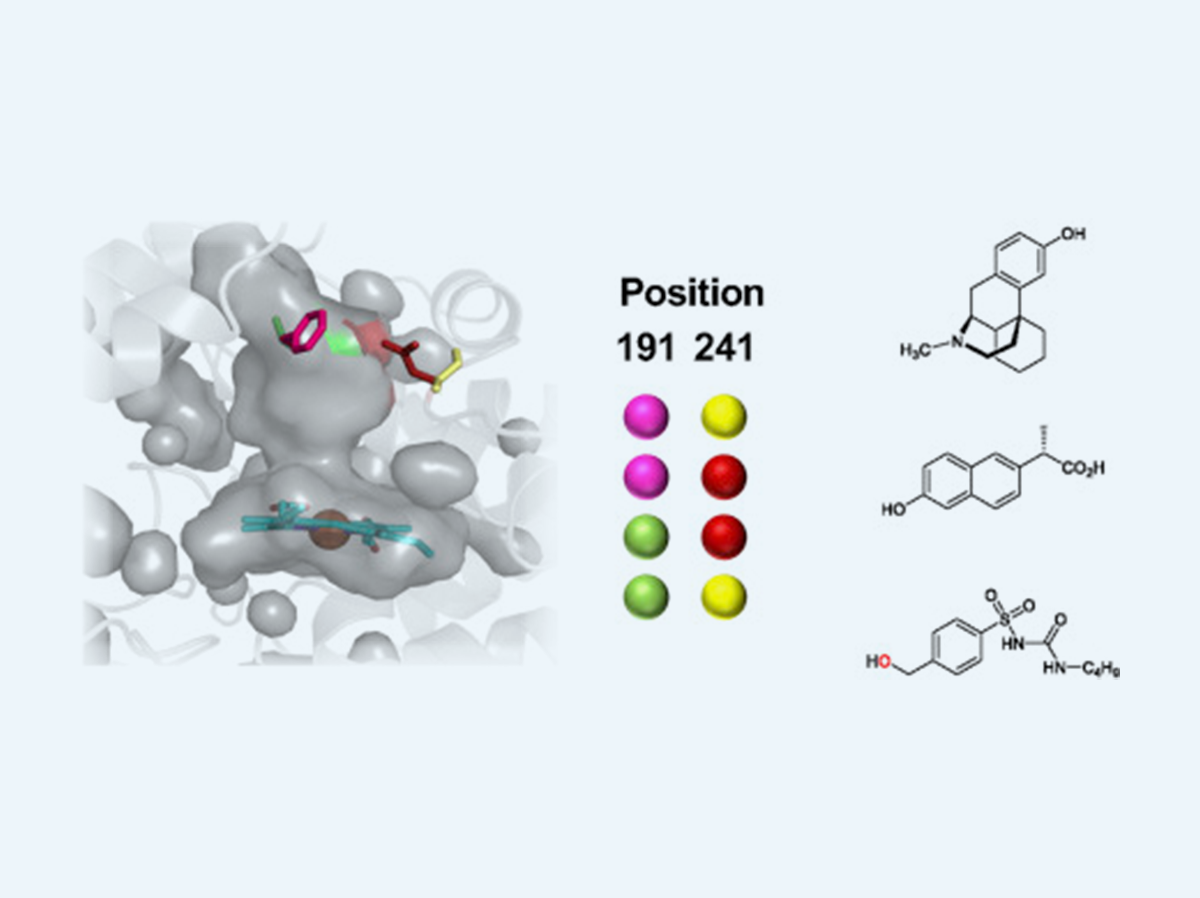
Benchmarking of laboratory evolved unspecific peroxygenases for the synthesis of human drug metabolites.
Gomez de Santos, P., Cervantes, F.V., Tieves, F., Plou, F.J., Hollmann, F. and Alcalde, M. (2019).
Tetrahedron 75: 1827-1831. https://pubs.acs.org/doi/10.1021/acssynbio.8b00509.